Medication Inventory Application
App for tracking medications, dispense history, and predicting which medications to buy for Main Street Family Medicine, PLLC.
In my role at Main Street Family Medicine, I created an application for our nurses to keep better track of our medications stored in-house. The app also features an option to predict which medicaions would be the best to re-order at any given time. This is based on our current inventory, dispense history, and other data concerning our current patient base. The prediction feature saves our nurses time it would otherwise take them to manually determine which medications they need to order.
Below includes a few screenshots of the app pages and the code I used to create the prediction model.
When a user navigates to the web application, they are first prompted to enter the password (as seen below):
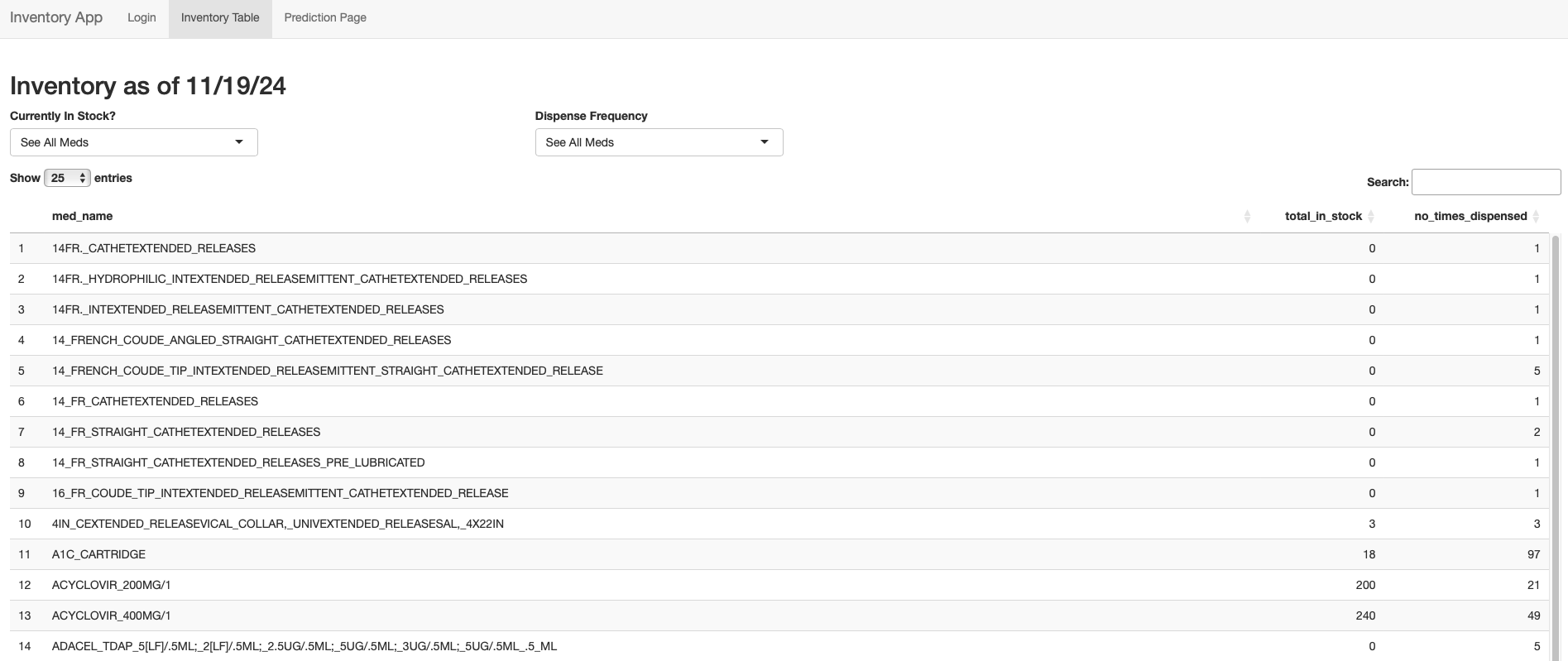
With the correct password, the app automatically navigates to the second page, titled “Inventory Table.” The table reflects our current medication inventory along with how many times each med has been dispensed in practice history. Dispense history is an important number to have handy, as it influences our nurses’ decisions on when or if they should reorder a certain medication. The table is searchable (see right-side search bar) and can be filtered by the number of medications in stock and/or the number of times each med has been dispense (see arrows next to table column names). These functions improve the usability of the app and make it more efficient for nurses to filter information when making ordering decisions.
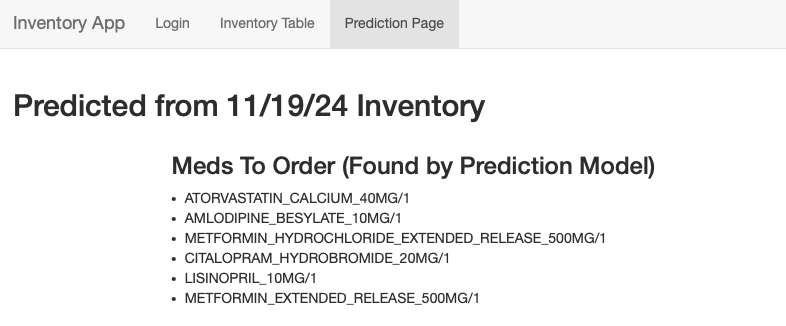
Finally, the last page (titled “Prediction Page”) lists the medications my model predicted we will need to order. This was based on current inventory, dispense history, previous order history, and various current patient demographic data. I included the code I used to create my model below. Also included on this page (though not in the screenshot for visability purposes) are lists of which frequently dispensed meds we are completely (and almost) out of.
The below code showcases my model for determining which meds we need to order and a summary of this process. This model was made in Python and the app in R (app code not shown).
- Preprocess data (OneHotEncoder and StandardScaler for categorical and numeric data, respectively)
- Test/train split
- Create an ensemble made of various regression models and determine which had the lowest negative mean squared error
- Hyperparameterized best model (found from previous step)
- Cross validate
- Test
- Predict
# 1. Preprocess
#standard scale all except NDC
#set up for categorical
encoder = OneHotEncoder(handle_unknown='ignore') #was using ordinalencoder but problem was that there were NDCs in test that were not in training, giving me best value of nan in my model. changed to onehotencoder to prevent this
categorical_cols = ["NDC"]
#set up for numerical
std_scaler = StandardScaler()
all_cols = df_final.columns.tolist()
numeric_cols = [col for col in all_cols if col not in categorical_cols]
numeric_cols.remove('total_ordered') #remove the final output
#now use a column transformer to do both pre processors on their separate columns
transformer = ColumnTransformer(
transformers=[
('categories', encoder, categorical_cols),
('numeric', std_scaler, numeric_cols)
],
remainder='drop', verbose_feature_names_out=False)
#####################
# 2. Split
X = df_final.loc[:, df_final.columns != 'total_ordered']
y = df_final['total_ordered']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
#####################
# 3. Ensemble
#four models that might work well
clf1 = RandomForestRegressor(random_state=0)
clf2 = GradientBoostingRegressor()
clf3 = BaggingRegressor()
clf4 = SGDRegressor()
#ensemble
eclf = VotingRegressor(
estimators=[('rf', clf1), ('boost', clf2), ('bag', clf3), ('sgd',clf4)])
#find accuracies using cross validated scores
for clf, label in zip([clf1, clf2, clf3, clf4, eclf], ['Random Forest', 'Gradient Boost', 'Bagging', 'SGD', 'Ensemble']):
scores = cross_val_score(clf, X_train_trans, y_train, scoring='neg_mean_squared_error', cv=5)
print("MSE: %0.2f (+/- %0.2f) [%s]" % (scores.mean(), scores.std(), label)) #gradient boost was the best
#####################
# 4. Hyperparameterize
#pipeline
pipeline = Pipeline([('transformer', transformer),
('GradBoost', GradientBoostingRegressor())])
#find names of parameters
pipeline.get_params()
#don't need to hyperparameterize the transformer
parameters = {
'GradBoost__learning_rate': [0.01, 0.1, 0.2, 0.3],
'GradBoost__n_estimators': randint(1,1000),
'GradBoost__max_depth': randint(1,10),
'GradBoost__min_impurity_decrease': [0.0, 0.1, 0.2, 0.3],
}
n_iter_search = 5
random_search = RandomizedSearchCV(pipeline, param_distributions=parameters, n_iter=n_iter_search, n_jobs=-1, verbose=True)
#find best score
try:
random_search.fit(X_train, y_train)
# Find the best score
best_score = random_search.best_score_
print("Best score: %0.2f" % best_score)
except Exception as e:
print(f"An error occurred: {e}")
#Best negative MSE = 0.47- very good! (since y_train values range from 0 to 1400, see: y_train_counts = y_train.value_counts())
#####################
# 5. Cross validate
scores = cross_val_score(random_search, X_train, y_train, cv=5)
print(scores.mean(), '\t', scores.std())
# 0.3921064495878645 0.08086083233321788 - really happy with this
#find parameters used
random_search.best_estimator_
#learning rate = 0.01, max_depth = 2, n_estimators = 246
#####################
# 6. Test
#set up pipeline with above parameters
pipeline_test = Pipeline([('transformer', transformer),
('GradBoost', GradientBoostingRegressor(learning_rate=0.01, max_depth=2, n_estimators=246))])
#fit and predict
pipeline_test.fit(X_train, y_train)
y_pred = pipeline_test.predict(X_test)
#find neg mse
mse = mean_squared_error(y_test,y_pred)
neg_mse = -mse
#####################
# 7. Predict
y_pred_current = pipeline_test.predict(current_final)
#convert to df
predictions_df = pd.DataFrame(y_pred_current, columns=['Predicted_total_ordered'])
#match NDC and name to index
pred_counts = predictions_df['Predicted_total_ordered'].value_counts()
#print(pred_counts)
predictions_df_filter = predictions_df[predictions_df['Predicted_total_ordered']>1]
print(predictions_df_filter)
#use index to match
matching_rows = current_final.loc[predictions_df_filter.index]
#merge name with ndc
pred_names = pd.merge(names,matching_rows,on=["NDC"])
#keep only necessary names
cols_to_keep6 = ["Name","NDC"]
pred_names = pred_names[cols_to_keep6]
#drop duplicated NDC
pred_names = pred_names.drop_duplicates(subset=['NDC'])
pred_names = pred_names.drop_duplicates(subset=['Name'])
#drop NDC then export to csv for RStudio
pred_names = pred_names.drop(columns=["NDC"])
pred_names.to_csv("./prediction_names.csv")
NOTE: Thank you to the University of California for the stock medication image used for this project.